Resource
Collection of Scientific Knowledge Graphs (SciKGs) resources
Scientific knowledge graphs (SciKGs), as the core infrastructure of AI for Science, are reshaping the scientific research paradigm in unprecedented ways. They transform the vast, multi-source, and heterogeneous scientific knowledge (such as genes, compounds, materials, and reaction mechanisms) scattered across literature, databases, and experiments into semantic networks that machines can understand and reason with. This, in turn, provides structured knowledge support and decision-making basis for complex tasks such as drug discovery, new material design, and exploration of biological mechanisms.
This resource page is systematically organized and integrated based on the review paper 《Bridging Data and Discovery: A Survey on Knowledge Graphs in AI for Science》. This review is the first to comprehensively examine the complete technological chain and development trends of SciKGs from construction, application, to integration with Large Language Models (LLMs) from a global perspective across three fundamental disciplines: biology, chemistry, and materials. It is an authoritative reference for understanding the cutting-edge progress in this field.
We take this review as a blueprint and are committed to building an open and continuously updated scientific knowledge graph resource navigation station. Here, we not only list key public databases, core construction tools, and typical application cases, but also deeply deconstruct how the emerging paradigm of SciKG and LLM collaboration drives automated scientific discovery. All resources provide direct links and brief explanations, and are continuously maintained and updated in our GitHub repository(GitHub – HICAI-ZJU/SciKGs: A Survey on Knowledge Graphs in AI for Science) , aiming to lower the entry barrier to the field, help researchers quickly locate the required resources, and promote AI driven scientific innovation.
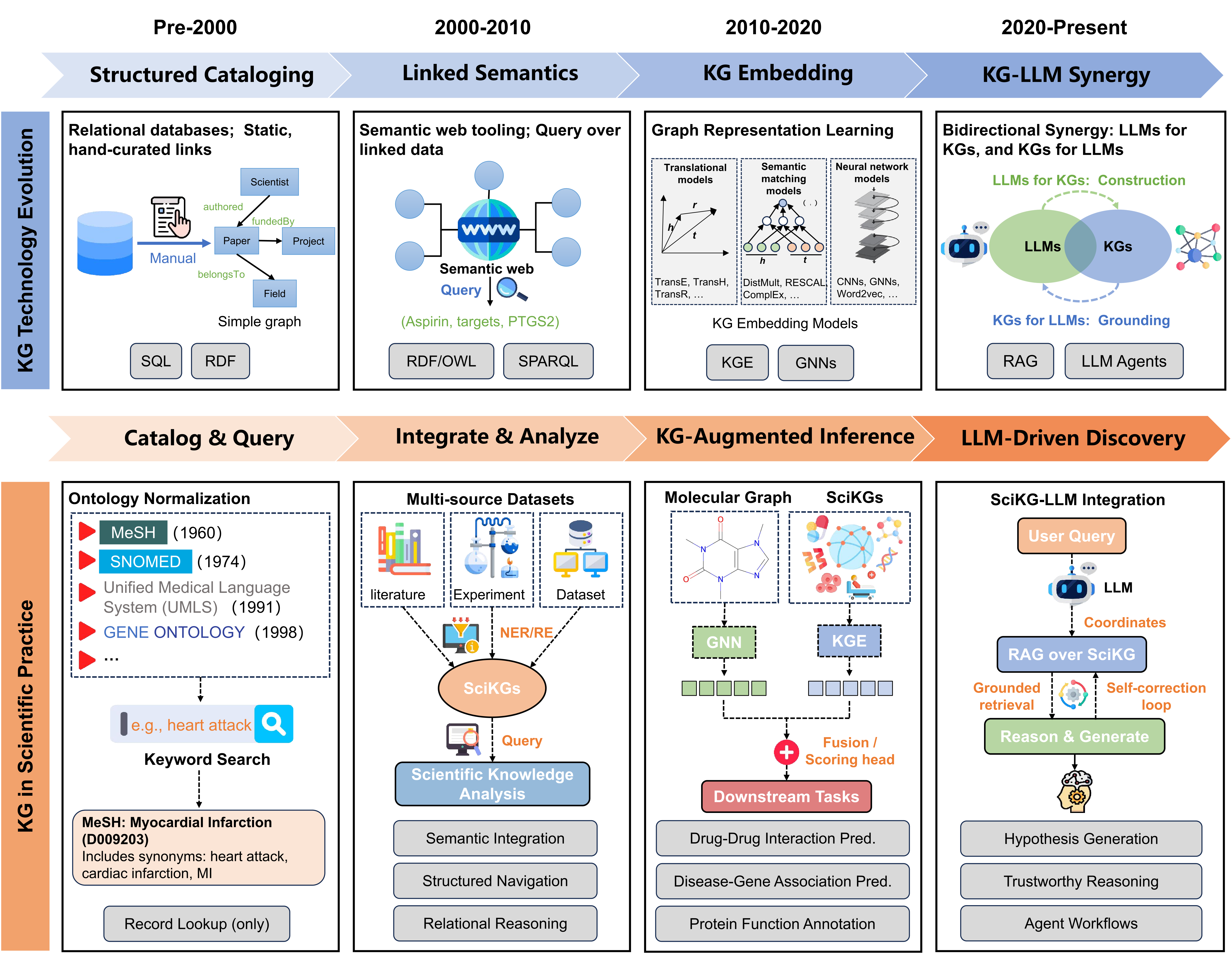
Figure 1: The evolution of scientific knowledge graph technology and the transformation of scientific practice.
- 🔬SciKG commonly used public databases: authoritative structured data sources covering the fields of biology, chemistry, and materials, which are the cornerstone of building high-quality SciKG.
- ⚙️ ScIKG construction and maintenance tool: a software and platform used for tasks such as knowledge extraction, ontology alignment, graph fusion, dynamic updates, and inference completion.
- 🚀SciKG core application scenario classification: Focusing on four major application scenarios: drug development and optimization, omics interpretation and analysis, chemical reactions and synthesis, and material design and synthesis, a total of 160+practical application cases have been summarized for 16 key tasks such as drug repositioning and reaction prediction.
- 🔄SciKG-LLM collaborative resources: frameworks, models, and application cases that support bidirectional empowerment of both (such as knowledge grounding, semantic reasoning, hypothesis generation).
We believe that structured knowledge is the cornerstone of future scientific intelligence. This page and the GitHub repository behind it will serve as the starting point for community collaboration. All researchers are welcome to use, supplement, and correct it.
Citation
If you find these resources useful, please cite our paper:
@article{Ding_2025,
title={Bridging Data and Discovery: A Survey on Knowledge Graphs in AI for Science},
url={http://dx.doi.org/10.36227/techrxiv.176369442.22009541/v1},
DOI={10.36227/techrxiv.176369442.22009541/v1},
publisher={Institute of Electrical and Electronics Engineers (IEEE)},
author={Ding, Keyan and Zhu, Zhihui and Tang, Yuqi and Feng, Kehua and Zhuang, Xiang and Wang, Hongwei and Yang, Yi and Du, Huifang and Ni, Zhangkai and Wang, Shiqi and Fan, Xiaohui and Xing, Huabin and Bai, Lei and Liu, Qi and Wang, Haofen and Zhang, Qiang and Chen, Huajun},
year={2025},
month=nov }
Drug Development and Optimization
| Year | Title | KG Name | KG Type | Domain | Construction Method | Venue | Paper |
|---|---|---|---|---|---|---|---|
| 2025 | TarIKGC: A Target Identification Tool Using Semantics-Enhanced Knowledge Graph Completion with Application to CDK2 Inhibitor Discovery | biological activity KG | public KG | DTI prediction | Semi-automated | Journal of Medicinal Chemistry | Link |
| 2025 | A comprehensive large-scale biomedical knowledge graph for AI-powered data-driven biomedical research | iKraph | Multi-source KG | Drug repurposing and Hypothesis Generation | Semi-automated | Nature Machine Intelligence | Link |
| 2025 | VITAGRAPH: Building a Knowledge Graph for Biologically Relevant Learning Tasks | VITAGRAPH | public KG | Drug repurposing | Semi-automated | arXiv | Link |
| 2024 | A Foundation Model for Clinician-Centered Drug Repurposing | / | public KG | Drug repurposing | Semi-automated | Nature Medicine | Link |
| 2024 | Accurate and Interpretable Drug-Drug Interaction Prediction Enabled by Knowledge Subgraph Learning | / | public KG | DDI prediction | Automated | Nature Communication Medicine | Link |
| 2024 | Knowledge Enhanced Representation Learning for Drug discovery | MKG | Multi-source KG | DTI prediction and Virtual screening and drug discovery | Semi-automated | AAAI | Link |
| 2024 | An experimentally validated approach to automated biological evidence generation in drug discovery using knowledge graphs | Healx KG | public KG | Drug repurposing | Semi-automated | Nature Communications | Link |
| 2024 | DDI-GPT: Explainable Prediction of Drug-Drug Interactions using Large Language Models enhanced with Knowledge Graphs | iBKH | public KG | DDI prediction | Semi-automated | bioRxiv | Link |
| 2024 | MKG-FENN: A Multimodal Knowledge Graph Fused End-to-End Neural Network for Accurate Drug–Drug Interaction Prediction | MKG | Multi-source KG | DDI prediction | Automated | AAAI | Link |
| 2024 | TransFOL: A Logical Query Model for Complex Relational Reasoning in Drug-Drug Interaction | / | public KG | DDI prediction | Semi-automated | Journal of Biomedical and Health Informatics | Link |
| 2024 | KGRLFF: Detecting Drug-Drug Interactions Based on Knowledge Graph Representation Learning and Feature Fusion | / | public KG | DDI prediction | Semi-automated | TCBB | Link |
| 2024 | An effective framework for predicting drug–drug interactions based on molecular substructures and knowledge graph neural network | DKG (Drug knowledge graph) | public KG | DDI prediction | Semi-automated | Computers in Biology and Medicine | Link |
| 2024 | Medical knowledge graph question answering for drug‐drug interaction prediction based on multi‐hop machine reading comprehension | / | public KG | DDI prediction | Automated | CAAI Transactions on Intelligence Technology | Link |
| 2024 | Integrated Knowledge Graph and Drug Molecular Graph Fusion via Adversarial Networks for Drug–Drug Interaction Prediction | DrugBank | public KG | DDI prediction | Semi-automated | JCIM | Link |
| 2024 | KGE-UNIT: toward the unification of molecular interactions prediction based on knowledge graph and multi-task learning on drug discovery | / | Multi-source KG | DDI prediction, DTI prediction and Hypothesis Generation | Automated | Briefings in Bioinformatics | Link |
| 2023 | Biomedical Knowledge Graph Learning for Drug Repurposing by Extending Guilt-By Association to Multiple Layers | / | public KG | Drug repurposing | Semi-automated | Nature Communications | Link |
| 2023 | Evolution-strengthened knowledge graph enables predicting the targetability and druggability of genes | ESKG (Evolution-strengthened KG) | public KG | DTI prediction | Semi-automated | PNAS nexus | Link |
| 2023 | Drugomics: Knowledge Graph & AI to Construct Physicians' Brain Digital Twin to Prevent Drug Side-Effects and Patient Harm | Drugomics KG | Multi-source KG | Drug toxicity and adverse reactions | Semi-automated | Big Data Analytics | Link |
| 2023 | Molecular-evaluated and explainable drug repurposing for COVID-19 using ensemble knowledge graph embedding | / | Multi-source KG | Drug repurposing | Semi-automated | Scientific Reports | Link |
| 2023 | Toxicology knowledge graph for structural birth defects | ReproTox-KG | Multi-source KG | Drug toxicity and adverse reactions and Hypothesis Generation | Semi-automated | Communications medicine | Link |
| 2023 | NAFLDkb: A Knowledge Base and Platform for Drug Development against Nonalcoholic Fatty Liver Disease | NAFLDkb | Multi-source KG | Drug repurposing | Semi-automated | JCIM | Link |
| 2023 | Molecule generation toward target protein (SARS-CoV-2) using reinforcement learning-based graph neural network via knowledge graph | / | domain-specific KG | Virtual screening and drug discovery, DTI prediction, and Hypothesis Generation | Semi-automated | Network Modeling and Analysis in Health Informatics and Bioinformatics | Link |
| 2022 | e-TSN: an Interactive Visual Exploration Platform for Target-Disease Knowledge Mappling from Literature | e-TSN KG | literature-based KG | DTI prediction | Automated | Briefings in Bioinformatics | Link |
| 2022 | Attention-based knowledge graph representation learning for predicting drug-drug interactions | / | public KG | DDI prediction | Semi-automated | Briefings in Bioinformatics | Link |
| 2022 | Automating Predictive Toxicology Using ComptoxAI | ComptoxAI KG | public KG | Drug toxicity and adverse reactions | Semi-automated | Chemical Research in Toxicology | Link |
| 2022 | KG-MTL: Knowledge Graph Enhanced Multi-Task Learning for Molecular Interaction | DRKG | public KG | DTI prediction | Automated | IEEE | Link |
| 2021 | A Unified Drug-Target Interaction Prediction Framework Based on Knowledge Graph and Recommendation System | / | public KG | DTI prediction | Semi-automated | Nature Communications | Link |
| 2021 | Biological Insights Knowledge Graph: an integrated knowledge graph to support drug development | BIKG | Multi-source KG | DTI prediction and Drug repurposing | Semi-automated | bioRxiv | Link |
| 2021 | SumGNN: multi-typed drug interaction prediction via efficient knowledge graph summarization | / | public KG | DDI prediction | Semi-automated | Bioinformatics | Link |
| 2021 | Predicting Potential Drug Targets Using Tensor Factorisation and Knowledge Graph Embeddings | Hetionet | public KG | DTI prediction | Semi-automated | BIOKDD | Link |
| 2021 | Adverse Drug Reaction Discovery Using a Tumor-Biomarker Knowledge Graph | TBKG (Tumor-biomarker KG) | literature-based KG | Drug toxicity and adverse reactions | Semi-automated | Frontiers in Genetics | Link |
| 2021 | Investigating ADR mechanisms with Explainable AI: a feasibility study with knowledge graph mining | PGxLOD | Multi-source KG | Drug toxicity and adverse reactions | Semi-automated | BMC Medical Informatics and Decision Making | Link |
| 2020 | Discovering Protein Drug Targets Using Knowledge Graph Embeddings | a knowledge graph of biological entities related to both drugs and targets | public KG | DTI prediction | Automated | Bioinformatics | Link |
| 2020 | KGNN: Knowledge Graph Neural Network for Drug-Drug Interaction Prediction | / | public KG | DDI prediction | Semi-automated | IJCAI | Link |
| 2020 | Network-based prediction of drug–target interactions using an arbitrary-order proximity embedded deep forest | / | public KG | DTI prediction | Semi-automated | Bioinformatics | Link |
| 2019 | Evaluation of knowledge graph embedding approaches for drug-drug interaction prediction in realistic settings | / | public KG | DDI prediction | Semi-automated | BMC Bioinformatics | Link |
| 2019 | Drug-Drug Interaction Prediction Based on Knowledge Graph Embeddings and Convolutional-LSTM Network | / | public KG | DDI prediction | Semi-automated | arXiv | Link |
| 2019 | GAMENet: Graph Augmented MEmory Networks for Recommending Medication Combination | EHR&DDI Graph | domain-specific KG | Virtual screening and drug discovery | Automated | AAAI | Link |
| 2019 | Facilitating prediction of adverse drug reactions by using knowledge graphs and multi‐label learning models | Bio2RDF KG | public KG | Drug toxicity and adverse reactions | Automated | Briefings in Bioinformatics | Link |
| 2018 | Modeling polypharmacy side effects with graph convolutional networks | / | public KG | Drug toxicity and adverse reactions | Semi-automated | Bioinformatics | Link |
| 2018 | Neural networks for link prediction in realistic biomedical graphs: a multi-dimensional evaluation of graph embedding-based approaches | / | public KG | DTI prediction | Semi-automated | BMC Bioinformatics | Link |
| 2017 | A Network Integration Approach for Drug-Target Interaction Prediction and Computational Drug Repositioning from Heterogeneous Information | / | public KG | DTI prediction and Drug repurposing | Semi-automated | Nature Communications | Link |
| 2017 | Knowledge graph prediction of unknown adverse drug reactions and validation in electronic health records | / | public KG | Drug toxicity and adverse reactions | Semi-automated | Scientific Reports | Link |
| 2017 | Large-scale structural and textual similarity-based mining of knowledge graph to predict drug–drug interactions | / | public KG | DDI prediction | Semi-automated | Journal of Web Semantics | Link |
| 2017 | Deep mining heterogeneous networks of biomedical linked data to predict novel drug–target associations | LTN (Linked Tripartite Network) | public KG | DTI prediction | Semi-automated | Bioinformatics | Link |
Omics Interpretation and Analysis
| Year | Title | KG Name | KG Type | Domain | Construction Method | Venue | Paper |
|---|---|---|---|---|---|---|---|
| 2025 | A novel approach for target deconvolution from phenotype-based screening using knowledge graph | P53_HUMAN PPIKG | public KG | Proteomics research | Semi-automated | Scientific Reports | Link |
| 2025 | Unified Knowledge-Guided Molecular Graph Encoder with multimodal fusion and multi-task learning | Elemental KG and Biological KG | Multi-source KG | Proteomics research | Semi-automated | Neural Networks | Link |
| 2025 | PhenoKG: Knowledge Graph-Driven Gene Discovery and Patient Insights from Phenotypes Alone | PhenoKG | public KG | Genomics research | Semi-automated | arXiv | Link |
| 2024 | Petagraph: A large-scale unifying knowledge graph framework for integrating biomolecular and biomedical data | Petagraph | Multi-source KG | Genomics research | Semi-automated | Scientific Data | Link |
| 2024 | An ontology-based knowledge graph for representing interactions involving RNA molecules | RNA-KG | Multi-source KG | Transcriptomics research | Semi-automated | Scientific Data | Link |
| 2024 | Knowledge graph construction based on granulosa cells transcriptome from polycystic ovary syndrome with normoandrogen and hyperandrogen | causal KG | Multi-source KG | Transcriptomics research | Semi-automated | Journal of Ovarian Research | Link |
| 2024 | Multi-Modal Protein Knowledge Graph Construction and Applications (Student Abstract) | ProteinKG65 | Multi-source KG | Proteomics research | Semi-automated | AAAI | Link |
| 2024 | Bridging chemical structure and conceptual knowledge enables accurate prediction of compound-protein interaction | DRKG | public KG | Proteomics research | Manual | BMC Biology | Link |
| 2024 | Integration of chromosome locations and functional aspects of enhancers and topologically associating domains in knowledge graphs enables versatile queries about gene regulation | crm, crm2gene, crm2tfac, crm2phen, tad, human genes (after ampliation) graph | public KG | Genomics research | Semi-automated | Nucleic Acids Research | Link |
| 2024 | Identifying compound-protein interactions with knowledge graph embedding of perturbation transcriptomics | / | public KG | Proteomics research | Semi-automated | Cell Genomics | Link |
| 2023 | MMiKG: a knowledge graph-based platform for path mining of microbiota–mental diseases interactions | MMiKG | literature-based KG | Microbiome research | Manual | Briefings in Bioinformatics | Link |
| 2023 | Transporter proteins knowledge graph construction and its application in drug development | Transporter Proteins Knowledge Graph | public KG | Proteomics research | Semi-automated | Computational and Structural Biotechnology Journal | Link |
| 2023 | A Knowledge Graph Approach to Elucidate the Role of Organellar Pathways in Disease via Biomedical Reports | / | Multi-source KG | Proteomics research | Semi-automated | JoVE Journal of Biochemistry | Link |
| 2022 | Knowledge-graph-based cell-cell communication inference for spatially resolved transcriptomic data with SpaTalk | LRT-KG | public KG | Transcriptomics research | Semi-automated | Nature Communications | Link |
| 2022 | A knowledge graph to interpret clinical proteomics data | CKG (Clinical Knowledge graph) | Multi-source KG | Proteomics research | Semi-automated | Nature Biotechnology | Link |
| 2022 | Knowledge integration and decision support for accelerated discovery of antibiotic resistance genes | E. coli knowledge graph | public KG | Genomics research | Semi-automated | Nature Communications | Link |
| 2022 | Machine learning prediction and tau-based screening identifies potential Alzheimer's disease genes relevant to immunity | PKG (protein knowledge graph) | public KG | Proteomics research | Semi-automated | Communications Biology | Link |
| 2022 | OntoProtein: Protein Pretraining With Gene Ontology Embedding | ProteinKG25 | public KG | Proteomics research | Automated | ICLR | Link |
| 2022 | GenomicKB: a knowledge graph for the human genome | GenomicKB (Genomic Knowledgebase) | Multi-source KG | Genomics research | Semi-automated | Nucleic Acids Research | Link |
| 2022 | Creating and Exploiting the Intrinsically Disordered Protein Knowledge Graph (IDP-KG) | IDP-KG | public KG | Proteomics research | Automated | CEUR Workshop Proceedings | Link |
| 2022 | BioTAGME: A Comprehensive Platform for Biological Knowledge Network Analysis | BioTAGME KG | Multi-source KG | Multi-Omics research | Semi-automated | Frontiers in Genetics | Link |
| 2022 | Biomedical knowledge graph embeddings for personalized medicine: Predicting disease-gene associations | a biomedical KG for predicting disease-gene association | public KG | Genomics research | Semi-automated | Expert Systems | Link |
| 2022 | Identifying genes targeted by disease-associated non-coding SNPs with a protein knowledge graph | Protein KG | literature-based KG | Genomics research | Semi-automated | PLOS ONE | Link |
| 2022 | KG-MTL: Knowledge Graph Enhanced Multi-Task Learning for Molecular Interaction | DRKG | public KG | Proteomics research | Automated | IEEE | Link |
| 2021 | FORUM: building a Knowledge Graph from public databases and scientific literature to extract associations between chemicals and diseases | FORUM | Multi-source KG | Metabolomics research | Semi-automated | Bioinformatics | Link |
| 2020 | Exploring the Microbiota-Gut-Brain Axis for Mental Disorders with Knowledge Graphs | MiKG (Microbiota knowledge graph) | Multi-source KG | Microbiome research | Semi-automated | Journal of Artificial Intelligence for Medical Sciences | Link |
| 2020 | Metastatic Site Prediction in Breast Cancer using Omics Knowledge Graph and Pattern Mining with Kirchhoff's Law Traversal | Kirchhoff's KG | public KG | Multi-Omics research | Semi-automated | bioRxiv | Link |
| 2020 | Accurate prediction of kinase-substrate networks using knowledge graphs | a phosphorylation knowledge graph | public KG | Proteomics research | Automated | PLoS Computational Biology | Link |
| 2020 | An integrative knowledge graph for rare diseases, derived from the Genetic and Rare Diseases Information Center (GARD) | an integrative KG for rare diseases | Multi-source KG | Genomics research | Semi-automated | Journal of Biomedical Semantics | Link |
| 2019 | Predicting gene-disease associations from the heterogeneous network using graph embedding | / | public KG | Genomics research | Semi-automated | IEEE | Link |
| 2019 | GenomicsKG: A Knowledge Graph to Visualize Poly-Omics Data | GenomicsKG | public KG | Genomics research | Semi-automated | Journal of Advances in Health | Link |
| 2018 | Semantic Disease Gene Embeddings (SmuDGE): phenotype-based disease gene prioritization without phenotypes | / | public KG | Genomics research | Semi-automated | Bioinformatics | Link |
| 2018 | Heterogeneous network embedding for identifying symptom candidate genes | SDGNet&SDGPNet (two heterogeneous symptom-related networks) | public KG | Genomics research | Semi-automated | JAMIA | Link |
| 2018 | Network-based integration of multi-omics data for prioritizing cancer genes | / | public KG | Genomics research | Semi-automated | Bioinformatics | Link |
| 2016 | A knowledge-based approach for predicting gene–disease associations | / | public KG | Genomics research | Semi-automated | Bioinformatics | Link |
Chemical Reaction and Synthesis
| Year | Title | KG Name | KG Type | Domain | Construction Method | Venue | Paper |
|---|---|---|---|---|---|---|---|
| 2025 | Bi-level Contrastive Learning for Knowledge-Enhanced Molecule Representations | MolKG | public KG | Molecular property prediction | Semi-automated | AAAI | Link |
| 2025 | Automated Retrosynthesis Planning of Macromolecules Using Large Language Models and Knowledge Graphs | / | literature-based KG | Chemical synthesis pathway optimization | Automated | Macromolecular Rapid Communications | Link |
| 2025 | An Automated Approach for Domain-Specific Knowledge Graph Generation─Graph Measures and Characterization | / | literature-based KG | Chemical reaction prediction and Chemical Synthesis Pathway Optimization | Automated | JCIM | Link |
| 2024 | Large-Scale Knowledge Integration for Enhanced Molecular Property Prediction | ElementKG-CHEBI | Multi-source KG | Molecular property prediction | Semi-automated | Nesy | Link |
| 2024 | Self-Supervised Contrastive Molecular Representation Learning with a Chemical Synthesis Knowledge Graph | Chemical synthesis KG | public KG | Chemical reaction prediction | Semi-automated | JCIM | Link |
| 2023 | Knowledge graph-enhanced molecular contrastive learning with functional prompt | ElementKG | domain-specific KG | Molecular property prediction | Semi-automated | Nature Machine Intelligence | Link |
| 2023 | Marie and BERT─A Knowledge Graph Embedding Based Question Answering System for Chemistry | TWA KG (the World Avatar KG) and Wikidata chemistry KG | Dynamic KG | Chemical reaction prediction and Molecular property prediction | Semi-automated | ACS Omega | Link |
| 2022 | MKGE: Knowledge graph embedding with molecular structure information | KCCR and DeepDDI | public KG | Molecular property prediction | Semi-automated | Computational Biology and Chemistry | Link |
| 2022 | Prediction of Compound Synthesis Accessibility Based on Reaction Knowledge Graph | Reaction knowledge graph | public KG | Chemical reaction prediction, Molecular property prediction, and Chemical synthesis pathway optimization | Semi-automated | Molecules | Link |
| 2022 | Molecular Contrastive Learning with Chemical Element Knowledge Graph | Chemical element KG | domain-specific KG | Molecular property prediction | Semi-automated | AAAI | Link |
| 2022 | FAIR and Interactive Data Graphics from a Scientific Knowledge Graph | / | literature-based KG | Chemical synthesis pathway optimization, Chemical reaction prediction, and Molecular property prediction | Semi-automated | Scientific Data | Link |
| 2022 | From Platform to Knowledge Graph: Evolution of Laboratory Automation | The World Avatar KG | Dynamic KG | Chemical Synthesis Pathway Optimization | Semi-automated | JACS Au | Link |
| 2021 | Intelligent generation of optimal synthetic pathways based on knowledge graph inference and retrosynthetic predictions using reaction big data | Reaction knowledge graph | public KG | Chemical synthesis pathway optimization | Semi-automated | Journal of the Taiwan Institute of Chemical Engineers | Link |
| 2021 | Automated Calibration of a Poly(oxymethylene) Dimethyl Ether Oxidation Mechanism Using the Knowledge Graph Technology | JPS KG | Dynamic KG | Chemical reaction prediction and Chemical Synthesis Pathway Optimization | Semi-automated | JCIM | Link |
| 2021 | A graph-based network for predicting chemical reaction pathways in solid-state materials synthesis | / | domain-specific KG | Chemical reaction prediction | Automated | Nature Communications | Link |
| 2020 | Knowledge Graph Approach to Combustion Chemistry and Interoperability | JPS KG | literature-based KG | Chemical reaction prediction | Automated | ACS Omega | Link |
| 2020 | Multiscale Cross-Domain Thermochemical Knowledge-Graph | JPS KG | Dynamic KG | Chemical reaction prediction and Molecular property prediction | Automated | JCIM | Link |
| 2016 | Modelling Chemical Reasoning to Predict and Invent Reactions | / | public KG | Chemical reaction prediction | Semi-automated | Chemistry Europe | Link |
Materials Design and Discovery
| Year | Title | KG Name | KG Type | Domain | Construction Method | Venue | Paper |
|---|---|---|---|---|---|---|---|
| 2025 | Construction of a knowledge graph for framework material enabled by large language models and its application | KG-FM | literature-based KG | Material screening and optimization | Semi-automated | npj Computational Materials | Link |
| 2025 | High throughput screening of new piezoelectric materials using graph machine learning and knowledge graph approach | a simple KG encoding structural similarity between materials | public KG | Material screening and optimization | Semi-automated | Computational Materials Science | Link |
| 2024 | MatKG: An autonomously generated knowledge graph in Material Science | MatKG | literature-based KG | New material design | Semi-automated | Scientific Data | Link |
| 2024 | A materials terminology knowledge graph automatically constructed from text corpus | MGED-KG | literature-based KG | Material screening and optimization | Semi-automated | Scientific Data | Link |
| 2024 | Construction and Application of Materials Knowledge Graph in Multidisciplinary Materials Science via Large Language Model | MKG | literature-based KG and dynamic KG | New material design | Semi-automated | NeurIPS | Link |
| 2024 | Generative Retrieval-Augmented Ontologic Graph and Multiagent Strategies for Interpretive Large Language Model-Based Materials Design | Ontological KG | literature-based KG | New material design and Material performance prediction | Semi-automated | ACS Engineering Au | Link |
| 2024 | SciAgents: Automating Scientific Discovery Through Bioinspired Multi-Agent Intelligent Graph Reasoning | an ontological knowledge graph for biologically inspired materials | literature-based KG | New material design and Material performance prediction | Semi-automated | Advanced Materials | Link |
| 2024 | An ontology-based text mining dataset for extraction of process-structure-property entities | Materials mechanics ontology | literature-based KG | Material performance prediction | Semi-automated | Scientific Data | Link |
| 2024 | Material Property Prediction with Element Attribute Knowledge Graphs and Multimodal Representation Learning | Element KG | domain-specific KG | Material performance prediction | Semi-automated | arXiv | Link |
| 2024 | Knowledge graph-guided data-driven design of ultra-high-performance concrete (UHPC) with interpretability and physicochemical reaction discovery capability | UHPC KG | literature-based KG | Material screening and optimization | Manual | Construction and Building Materials | Link |
| 2023 | The materials experiment knowledge graph | MekG | domain-specific KG | Material performance prediction | Semi-automated | Digital Discovery | Link |
| 2023 | Revisiting Electrocatalyst Design by a Knowledge Graph of Cu-Based Catalysts for CO2 Reduction | Cu-Based Catalysts Knowledge Graph for CO2 Reduction | literature-based KG | New material design and Material performance prediction | Semi-automated | ACS Catalysis | Link |
| 2023 | Reinforcement learning-based knowledge graph reasoning for aluminum alloy applications | Aluminum alloy domain KG | public KG | Material performance prediction | Semi-automated | Computational Materials Science | Link |
| 2023 | Bridging the Semantic-Numerical Gap: A Numerical Reasoning Method of Cross-modal Knowledge Graph for Material Property Prediction | Cross-modal KG | Multi-source KG | Material performance prediction | Semi-automated | arXiv | Link |
| 2023 | Digital Twin-Based Fault Diagnosis Platform for Final Rolling Temperature in Hot Strip Production | / | domain-specific KG | Material screening and optimization | Semi-automated | Materials | Link |
| 2022 | Grain Knowledge Graph Representation Learning: A New Paradigm for Microstructure-Property Prediction | Grain KG | domain-specific KG | Material performance prediction | Semi-automated | Crystals | Link |
| 2022 | Automating Materials Exploration with a Semantic Knowledge Graph for Li-Ion Battery Cathodes | a semantic knowledge graph dedicated to LIB cathodes | literature-based KG | New material design | Semi-automated | AFM | Link |
| 2022 | High-Throughput Computing Assisted by Knowledge Graph to Study the Correlation between Microstructure and Mechanical Properties of 6XXX Aluminum Alloy | 6XXX Aluminum Alloy KG | domain-specific KG | Material screening and optimization and Material performance prediction | Semi-automated | Materials | Link |
| 2022 | FAIR and Interactive Data Graphics from a Scientific Knowledge Graph | / | literature-based KG | Material screening and optimization | Semi-automated | Scientific Data | Link |
| 2022 | Compound Knowledge Graph-Enabled AI Assistant for Accelerated Materials Discovery | CKG (Compound KG) | Multi-source KG | New material design | Semi-automated | Integrating Materials and Manufacturing Innovation | Link |
| 2021 | EBSD Grain Knowledge Graph Representation Learning for Material Structure-Property Prediction | EBSD Grain KG | domain-specific KG | Material performance prediction | Semi-automated | CCKS | Link |
| 2020 | propnet: A Knowledge Graph for Materials Science | propnet KG | domain-specific KG | Material performance prediction | Semi-automated | Matter | Link |
| 2020 | NanoMine: A Knowledge Graph for Nanocomposite Materials Science | NanoMine KG | domain-specific KG | New material design | Semi-automated | ISWC | Link |
| 2018 | Relation extraction with weakly supervised learning based on process-structure-property-performance reciprocity | PSPP KG (Process-Structure-Property-Performance) | literature-based KG | New material design | Semi-automated | Science and Technology of Advanced Materials | Link |
