Applications
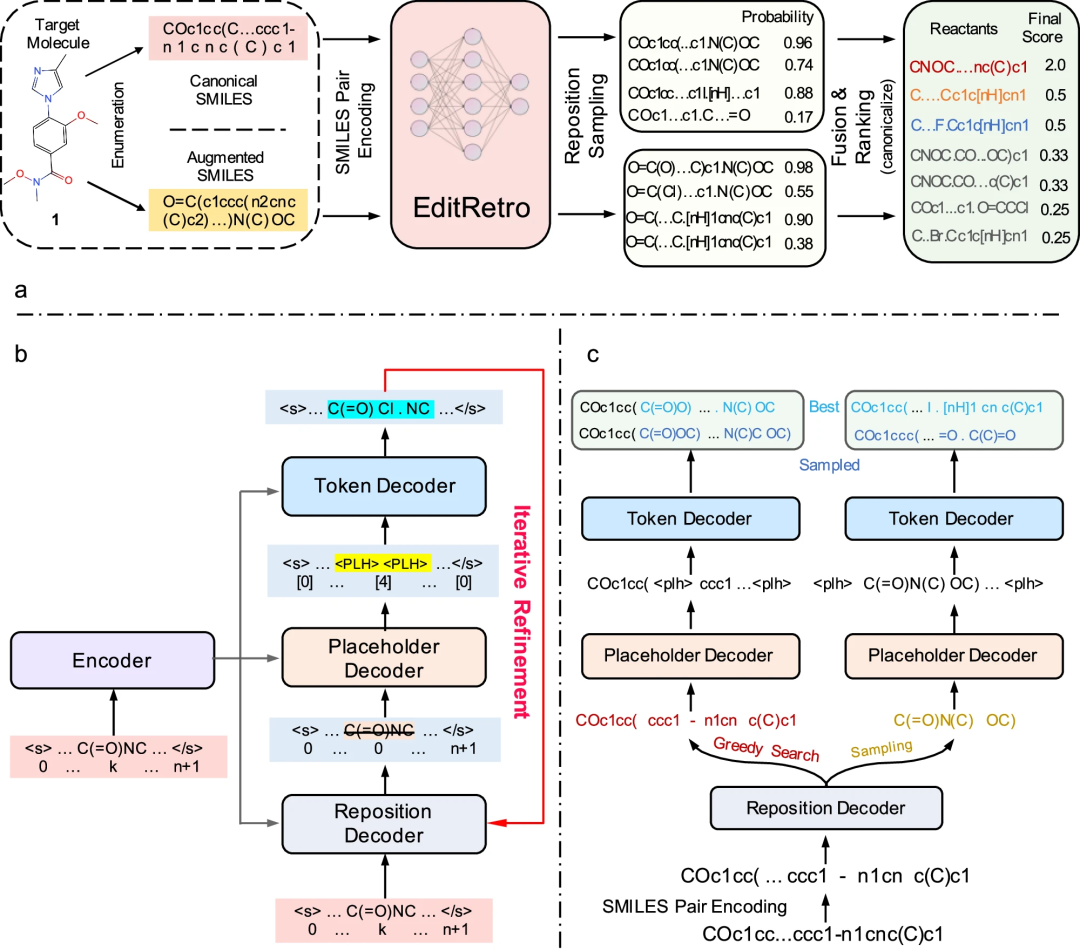
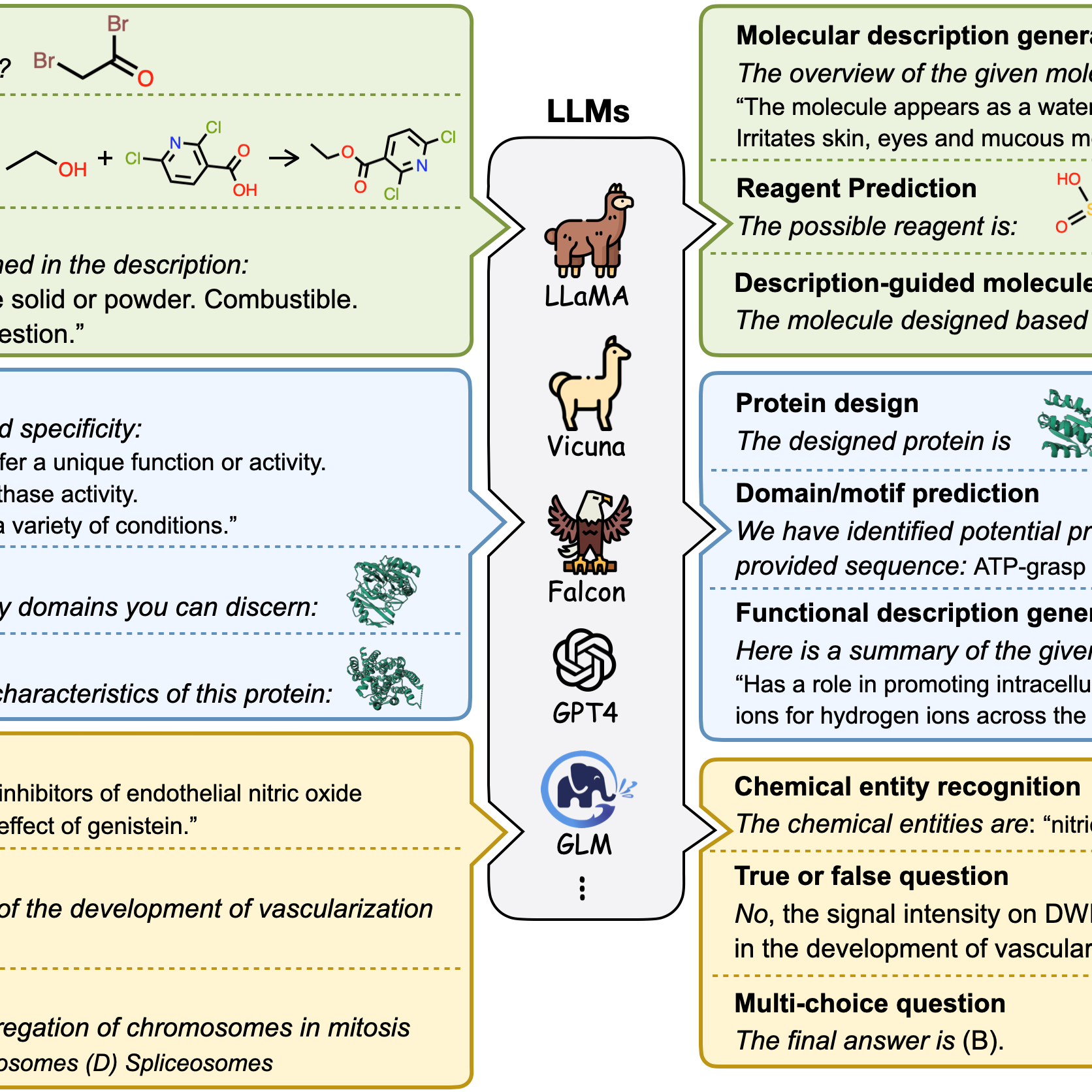
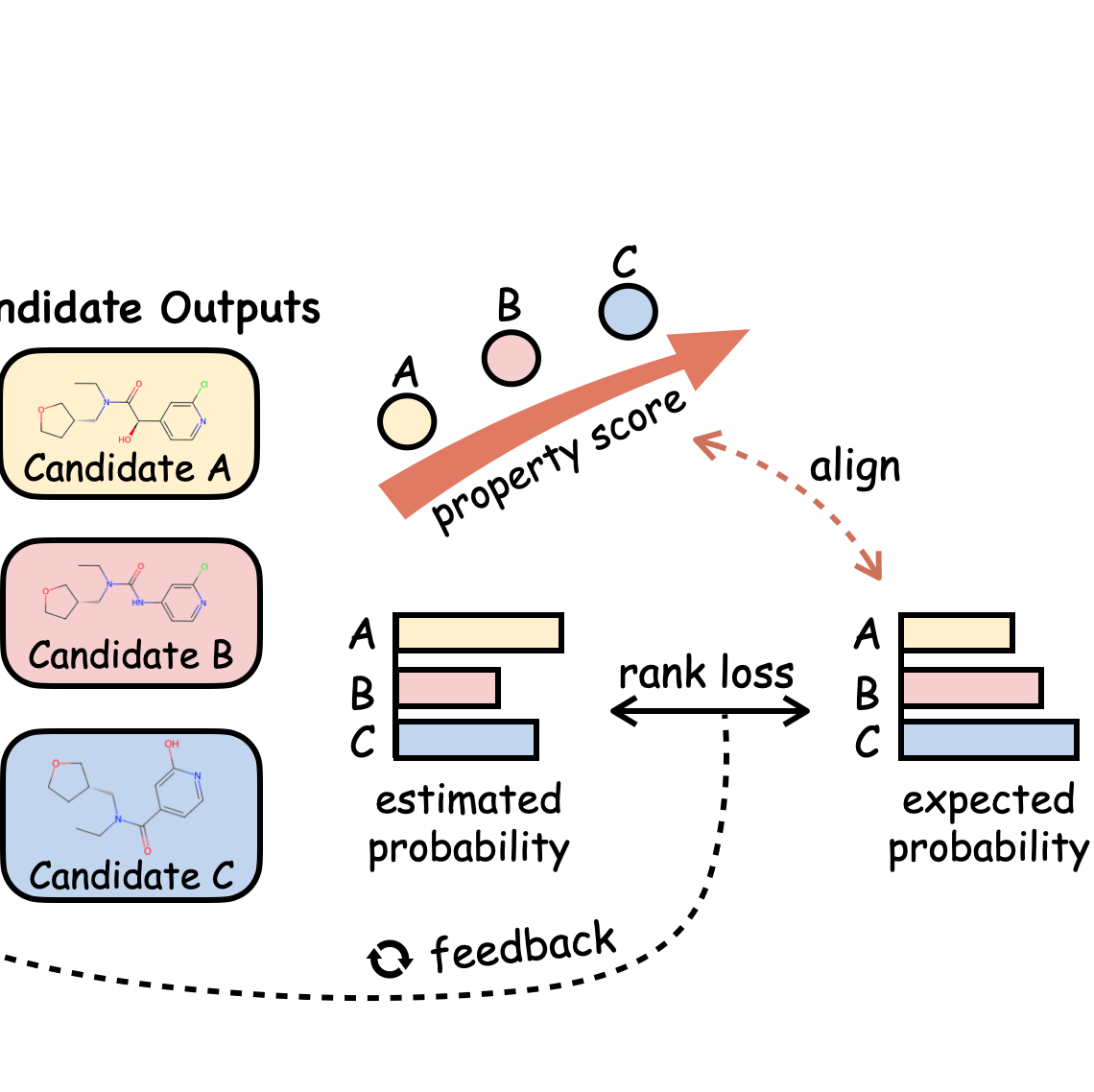
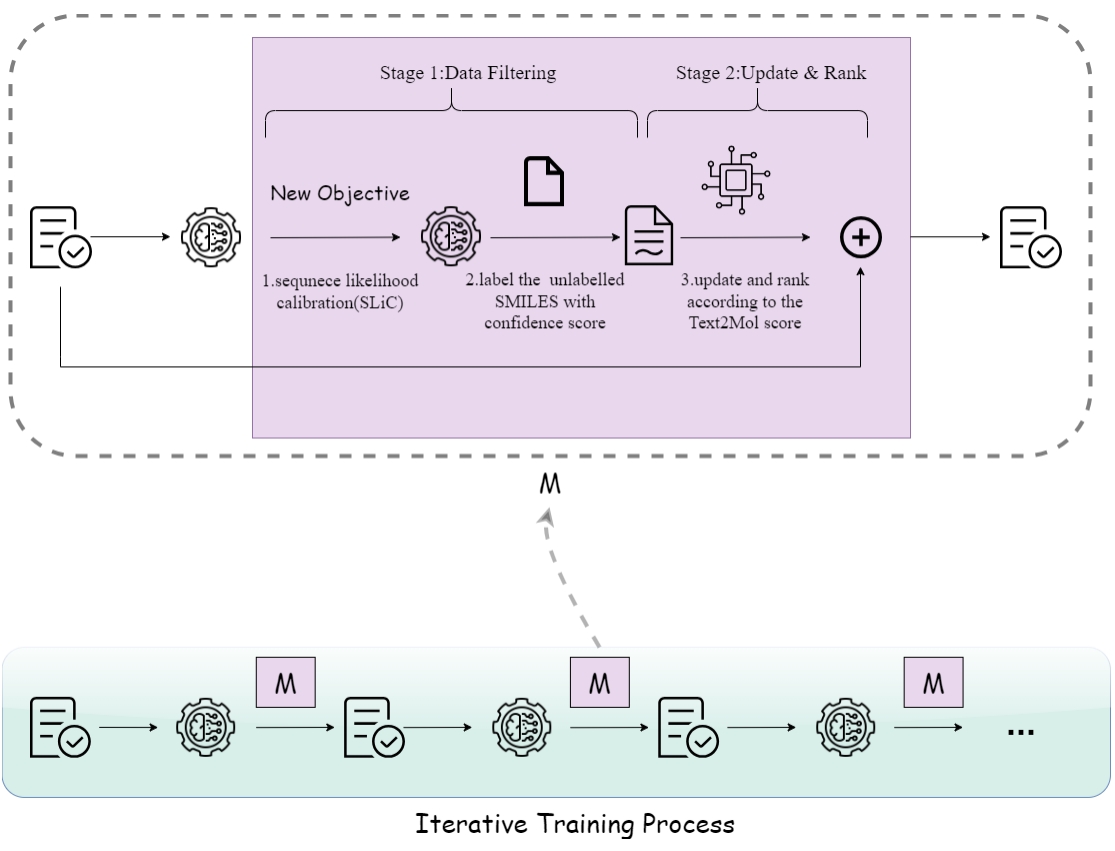
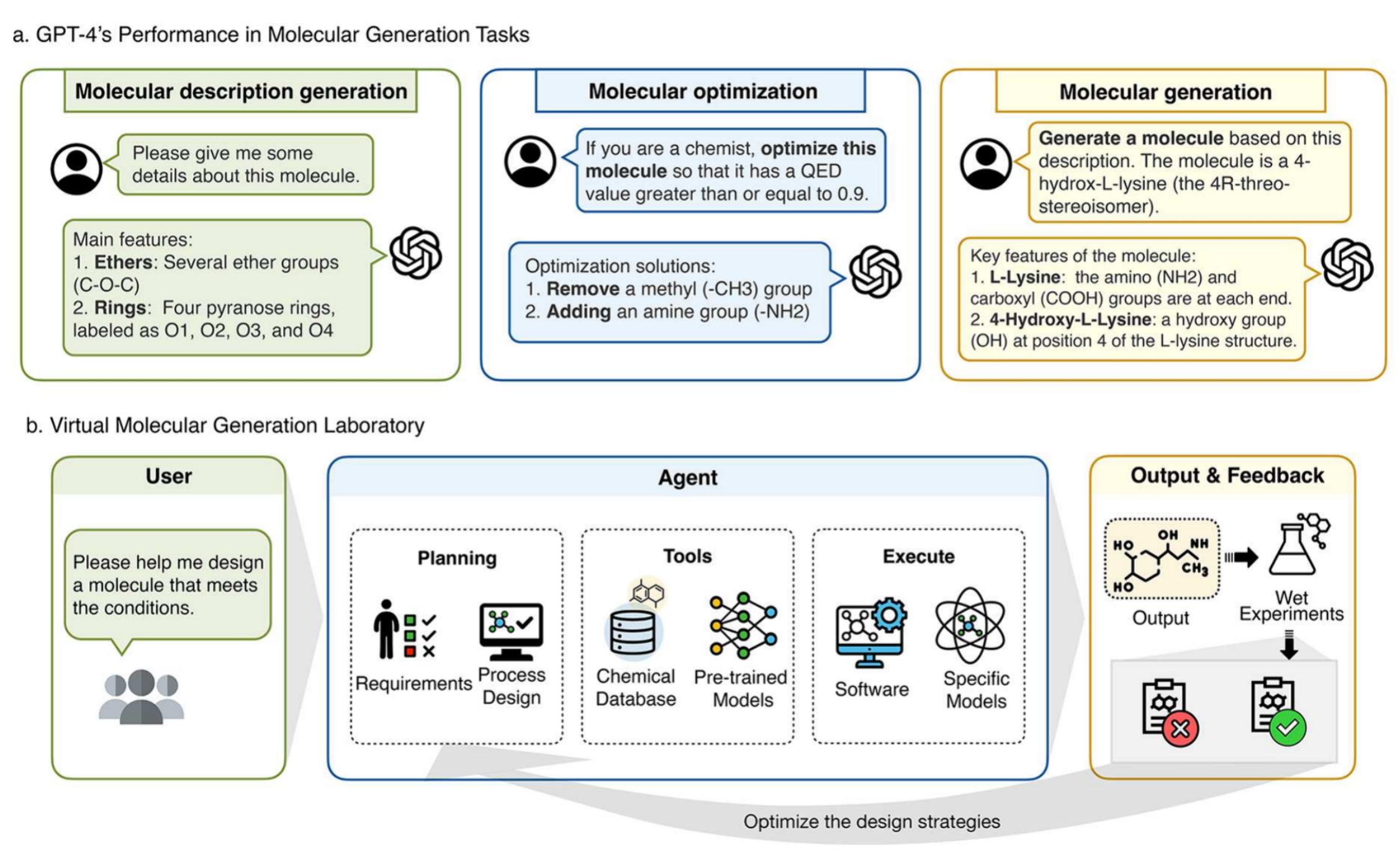
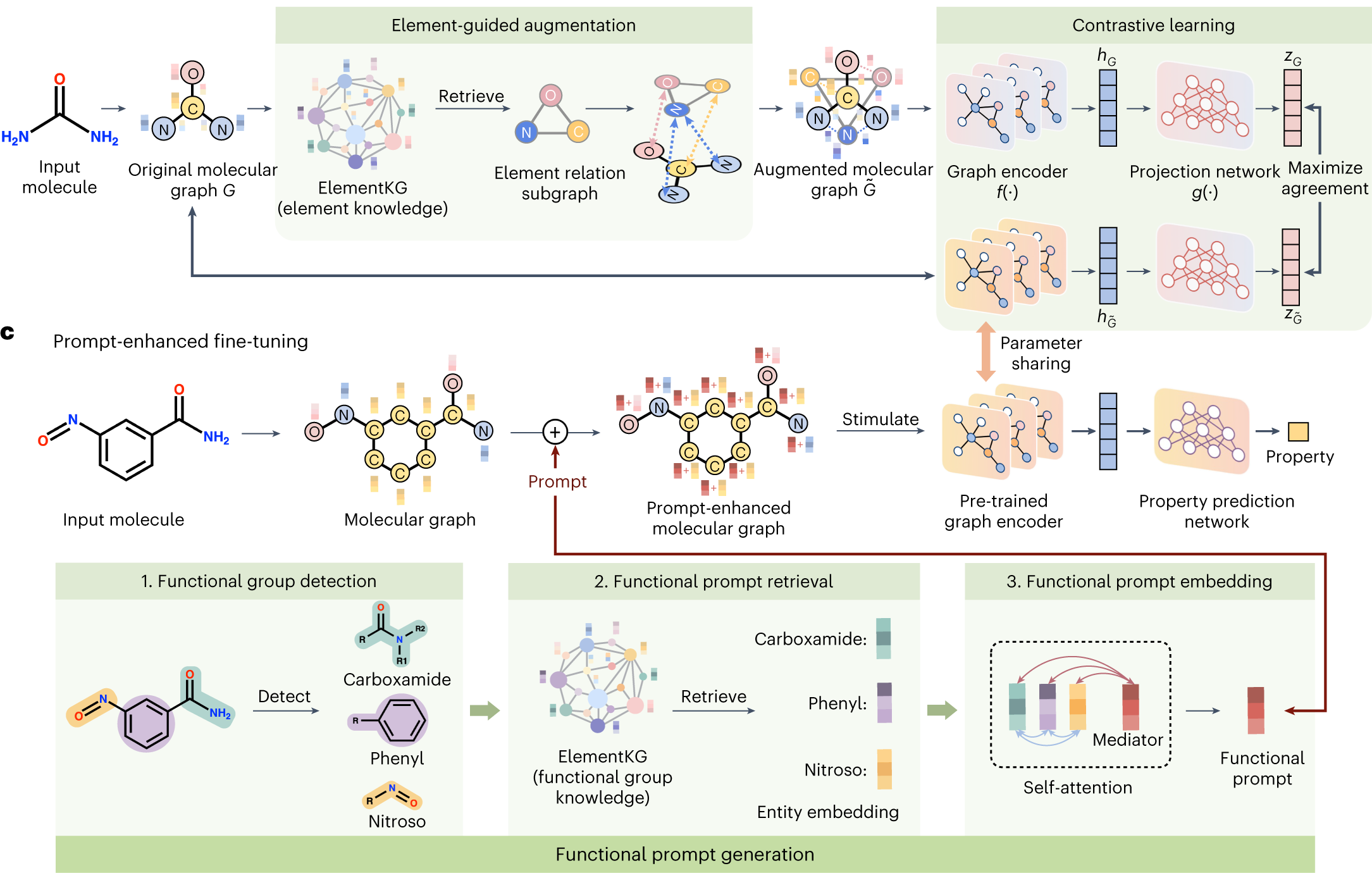
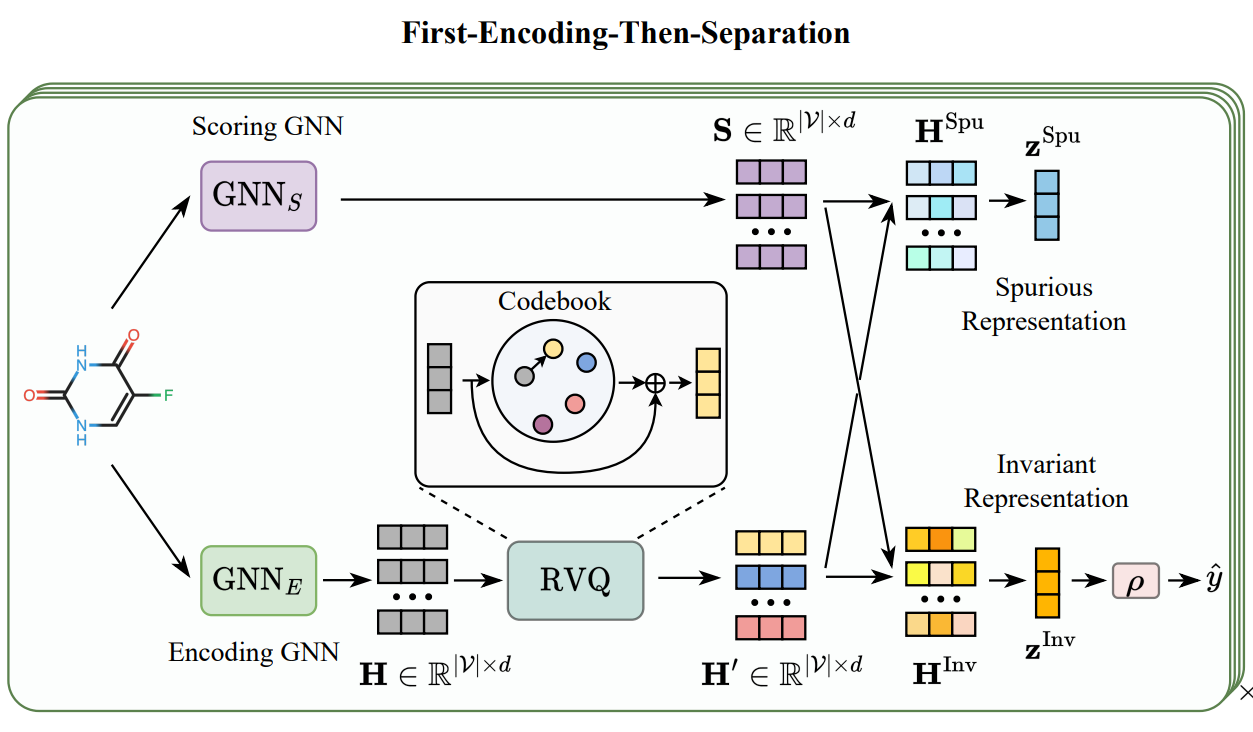
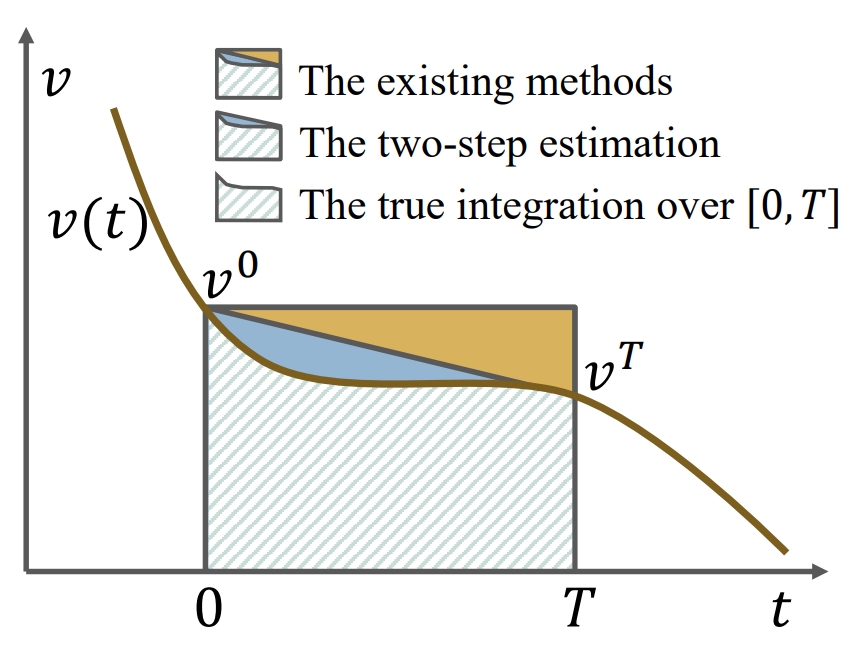
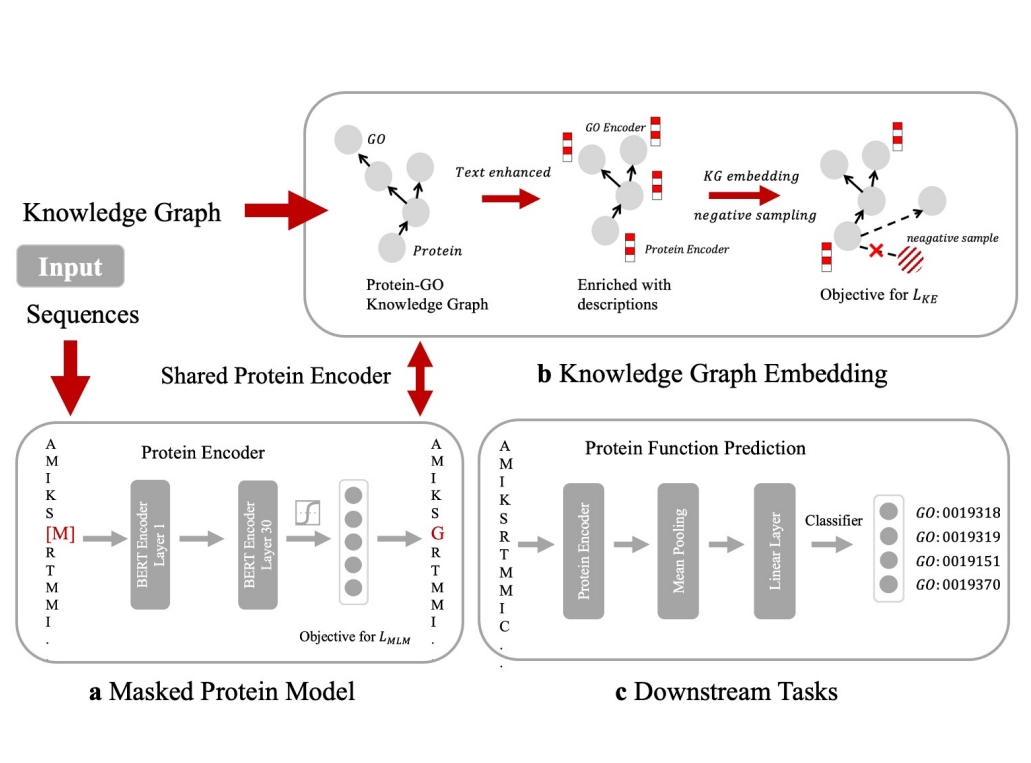
Integrating protein language models and automatic biofoundry for enhanced protein evolution
Nature Communication, 2025
Qiang Zhang, Wanyi Chen, Ming Qin, Yuhao Wang, Zhongji Pu, Keyan Ding, Yuyue Liu, Qunfeng Zhang, Dongfang Li, Xinjia Li, Yu Zhao, Jianhua Yao, Lei Huang, Jianping Wu, Lirong Yang, Huajun Chen & Haoran Yu
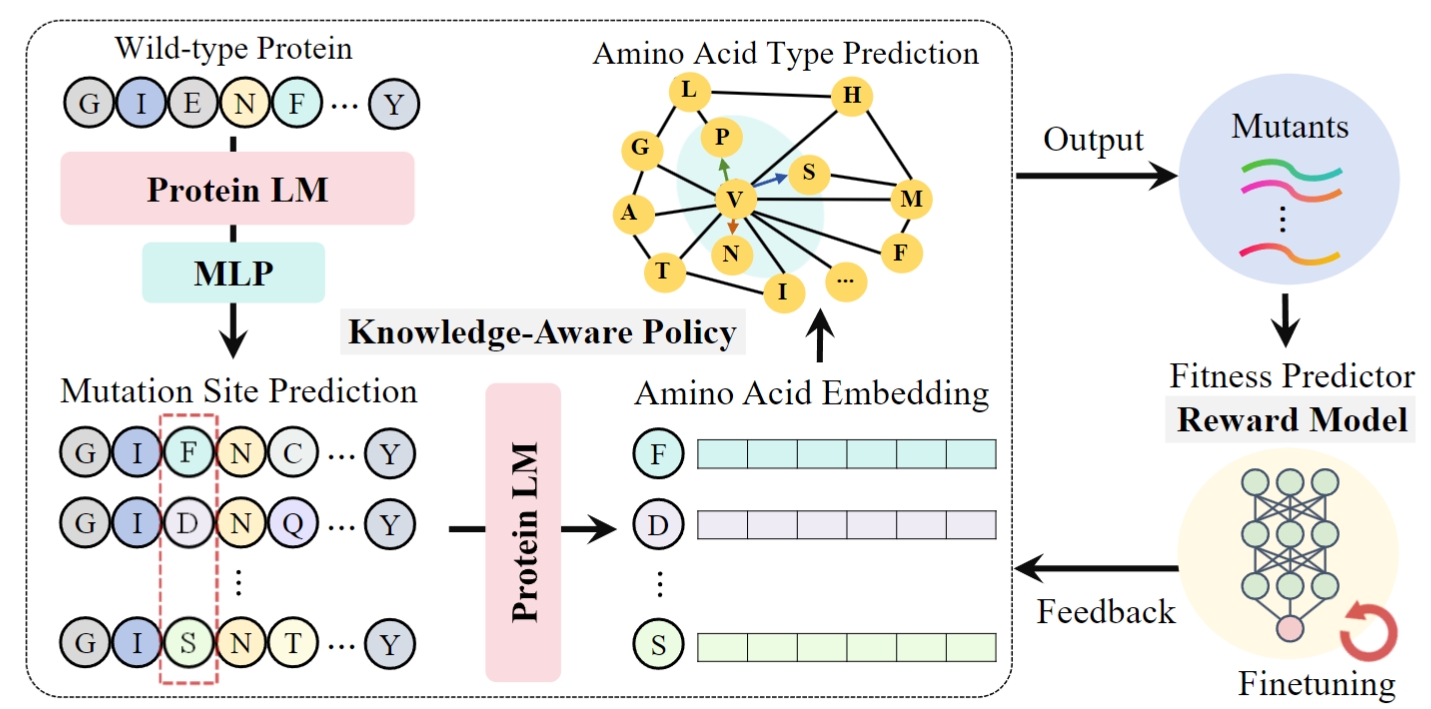
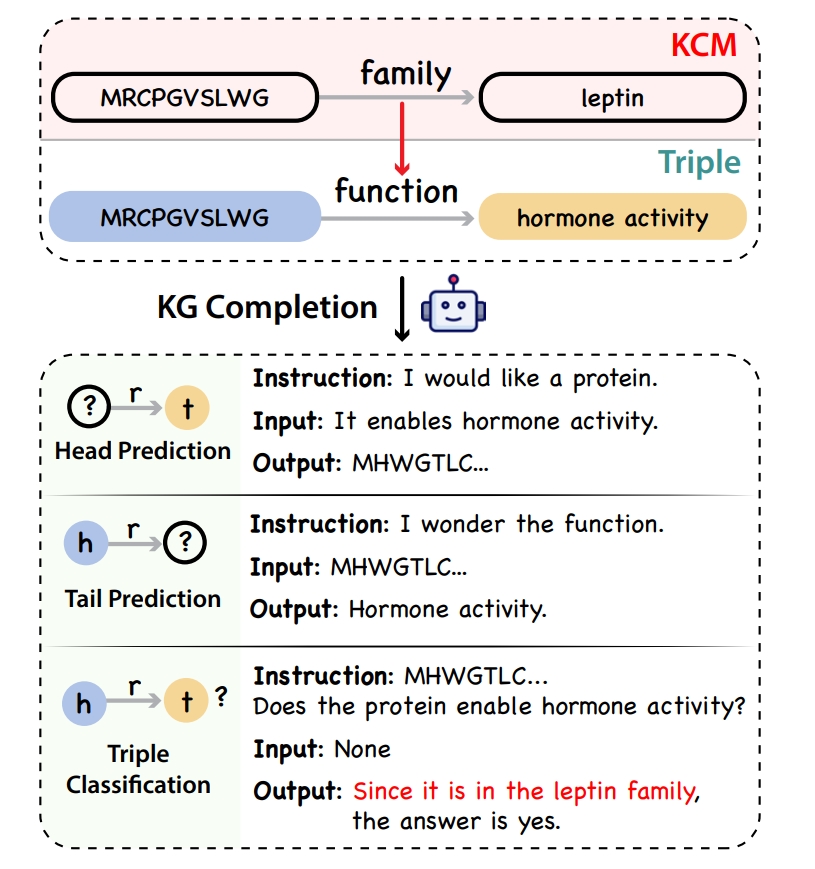
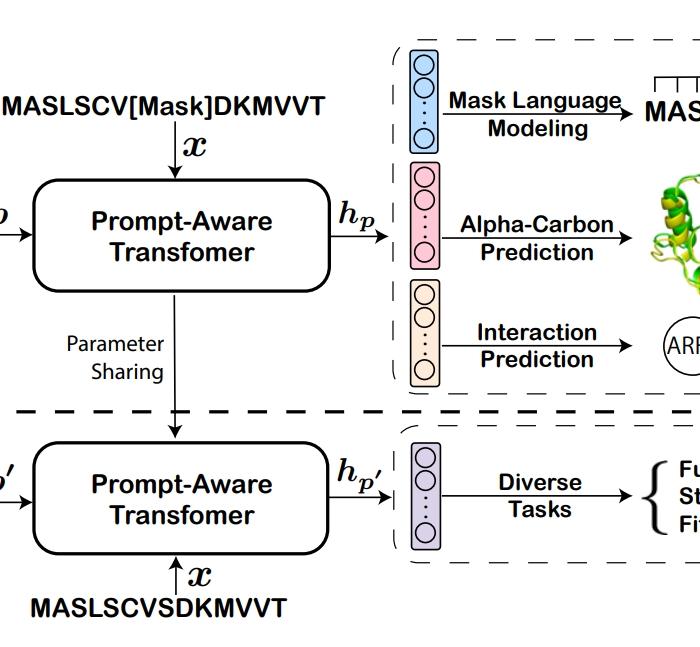
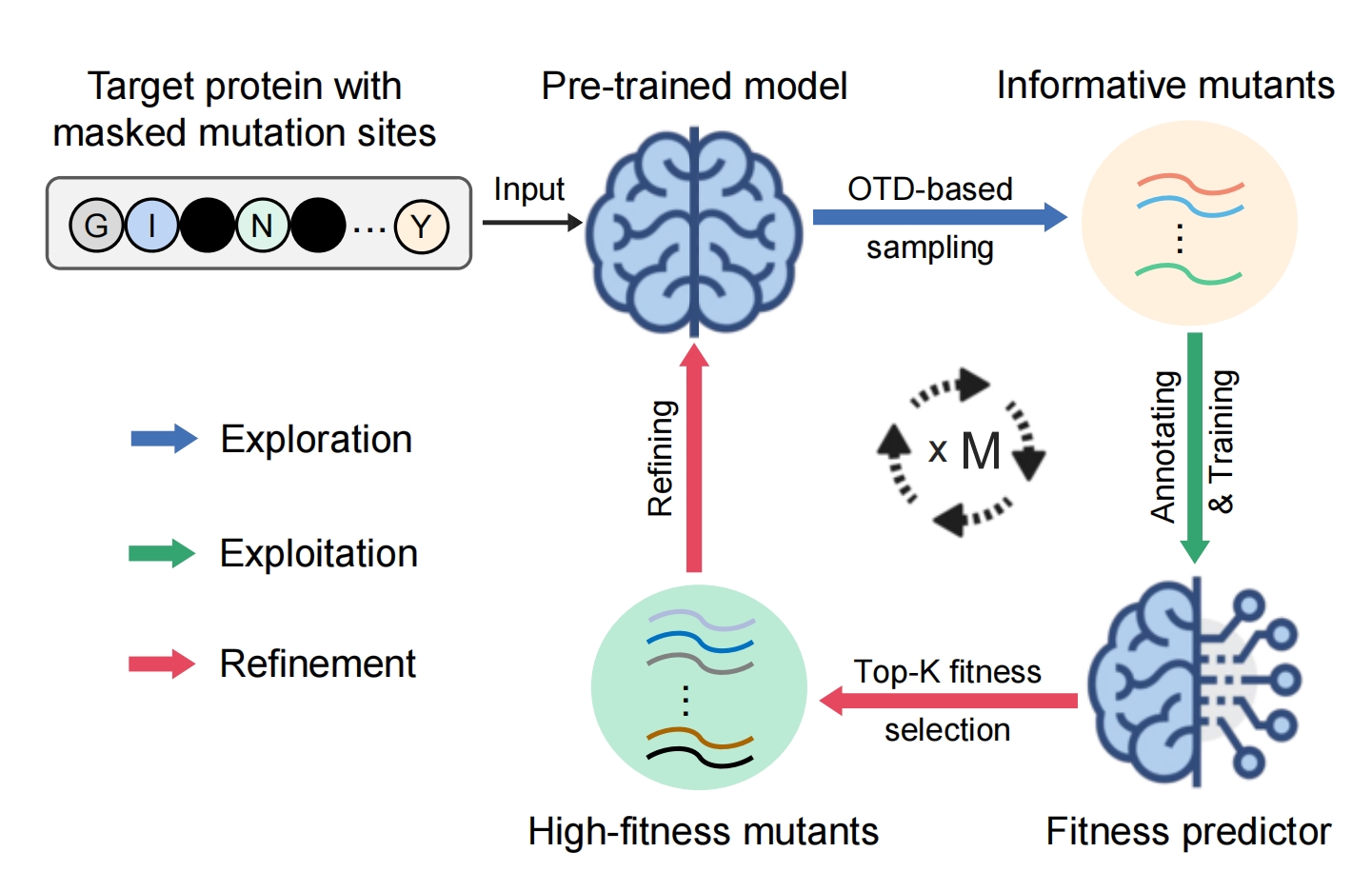
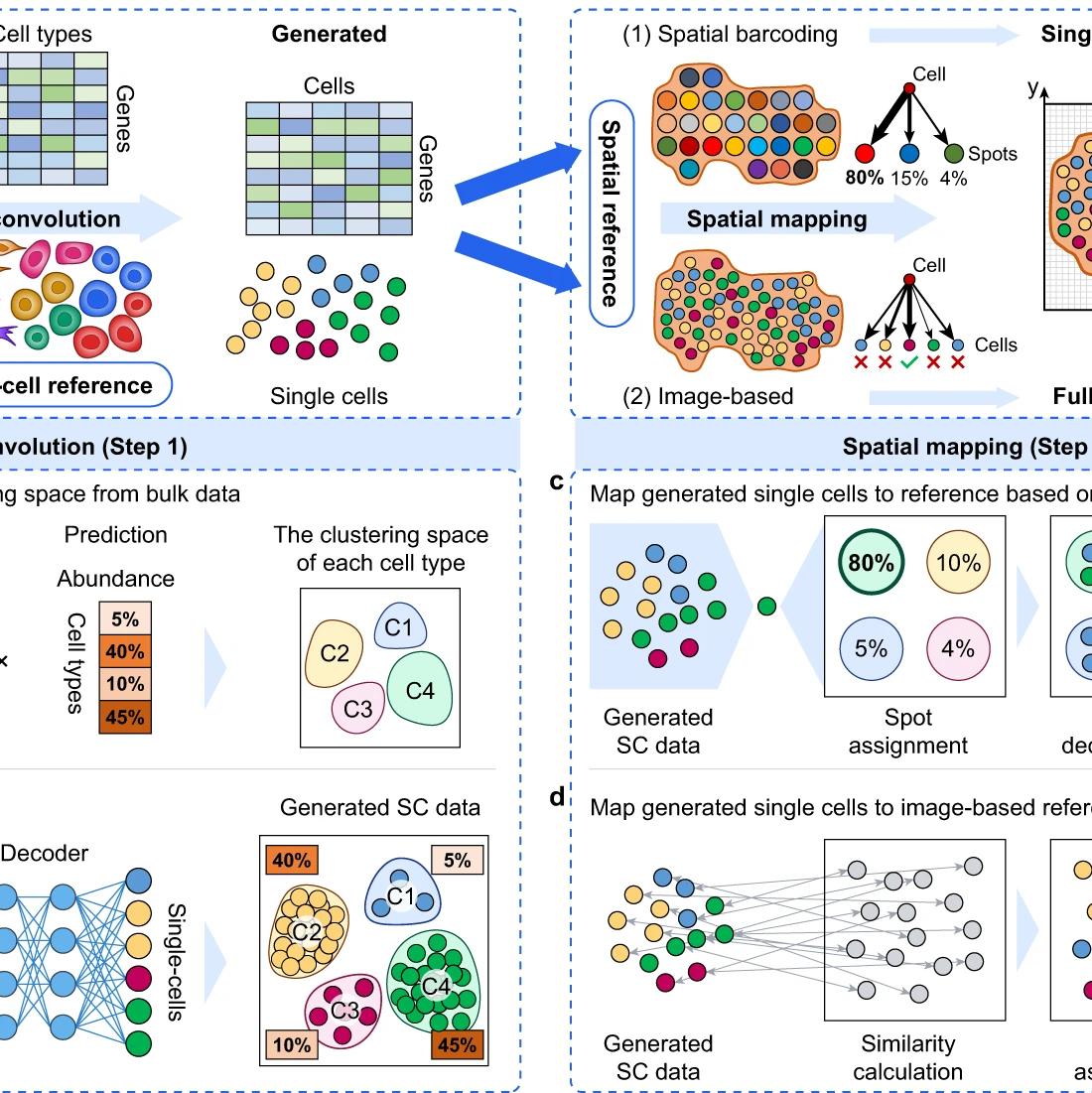
De Novo Analysis of Bulk RNA-seq Data at Spatially Resolved Single-cell Resolution
Nature Communications, 2022
Jie Liao , Jingyang Qian , Yin Fang , Zhuo Chen , Xiang Zhuang , Ningyu Zhang , Xin Shao , Yining Hu , Penghui Yang , Junyun Cheng , Yang Hu , Lingqi Yu , Haihong Yang , Jinlu Zhang , Xiaoyan Lu , Li Shao , Dan Wu , Yue Gao , Huajun Chen*, Xiaohui Fan*
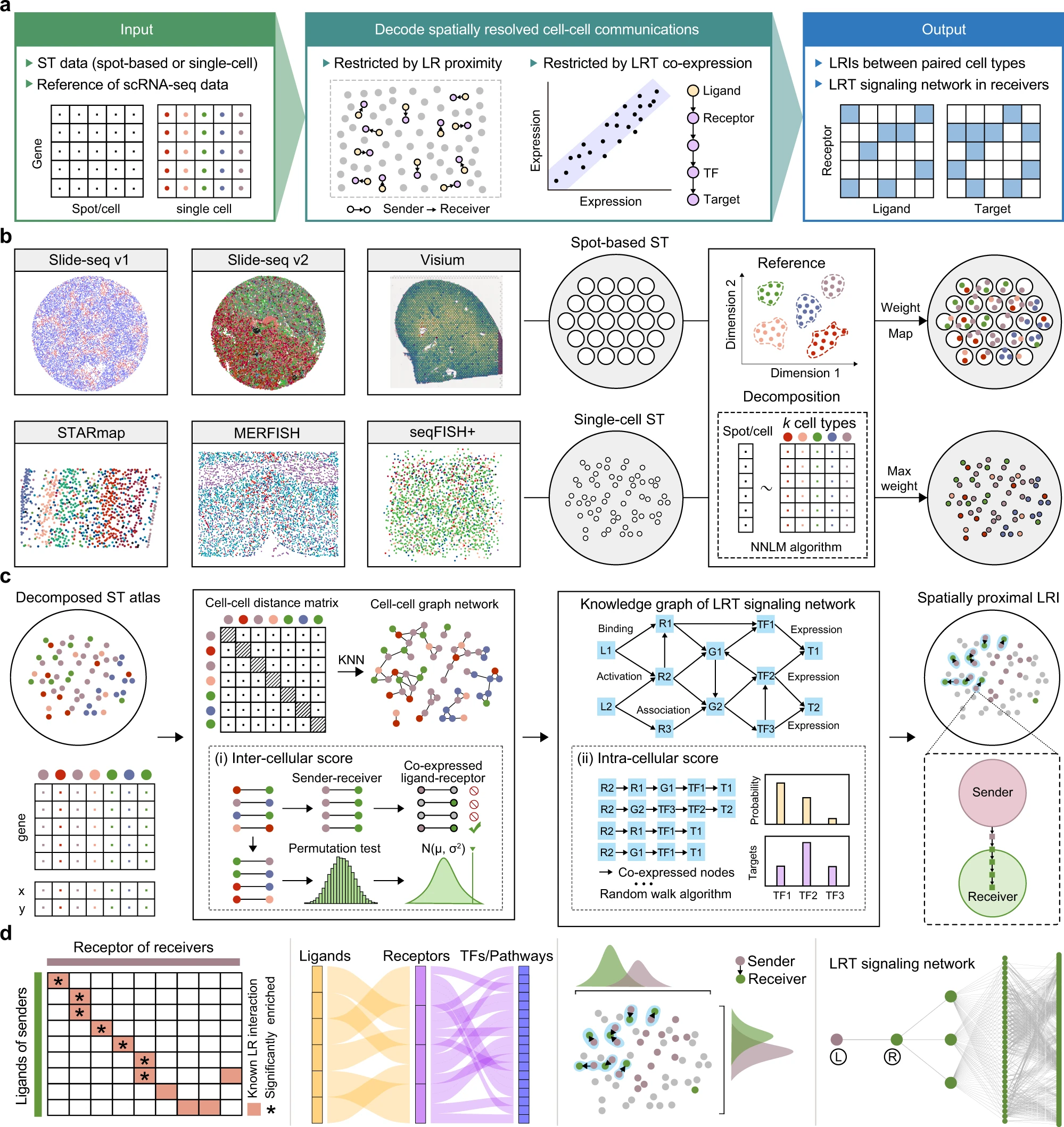
Knowledge-graph-based Cell-cell Communication Inference for Spatially Resolved Transcriptomic Data with SpaTalk
Nature Communications, 2022
Xin Shao, Chengyu Li, Haihong Yang, Xiaoyan Lu, Jie Liao, Jingyang Qian, Kai Wang, Junyun Cheng, Penghui Yang, Huajun Chen*, Xiao Xu*, Xiaohui Fan*